Accelerating Protein Structure Prediction: The xTrimo Multimer Breakthrough
Written on
Chapter 1: Introduction to xTrimo Multimer
The Colossal-AI team, in collaboration with BioMap, has recently launched an open-source tool called xTrimo Multimer, designed for predicting protein monomer and multimer structures. This groundbreaking solution can perform simultaneous predictions, enhancing efficiency by up to elevenfold!
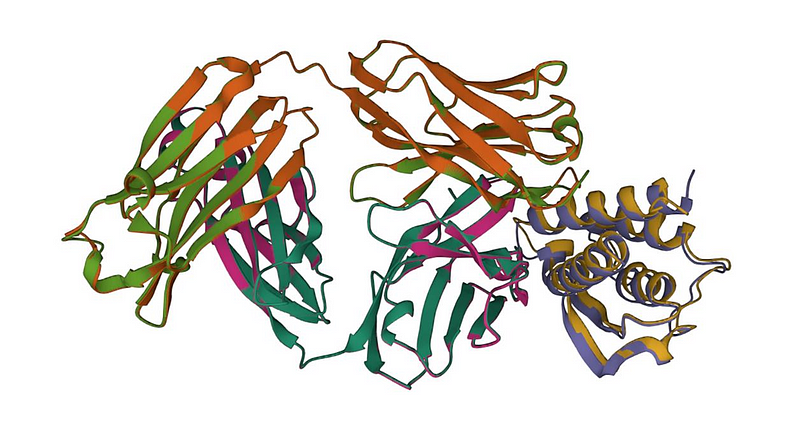
The driving force behind this innovation is Colossal-AI, a robust deep learning framework that simplifies the training of large AI models for both the community and industry. By leveraging the advanced training techniques and optimizations from Colossal-AI, the process of predicting protein structures can be significantly expedited, ultimately lowering both time and costs involved in model training and inference. As a notable application of Colossal-AI in the pharmaceutical sector, xTrimo Multimer promises to accelerate model design and development for protein structure prediction, paving the way for major advancements in AI applications within healthcare and bioinformatics.
Colossal-AI also stands out as an accessible deep learning system that helps organizations optimize AI deployments while minimizing expenses. Since its open-source release, Colossal-AI has frequently ranked among the top trending projects on GitHub and Papers With Code, gaining significant popularity with projects amassing up to 10,000 stars. The platform continues to expand its AI solutions across various industries, showing remarkable potential in areas such as medicine, self-driving technology, cloud services, retail, and semiconductor manufacturing. Recently, Colossal-AI partnered with BioMap to introduce a cost-effective approach to protein structure prediction, aiding healthcare professionals and pharmaceutical firms in diagnostics and pioneering new drug research.
Section 1.1: The Importance of Protein Structure Prediction
Understanding protein structure is crucial in structural biology, as it informs gene translation and protein functionality. However, the intricate multi-level structures and complex interactions among proteins present significant challenges in accurately predicting their three-dimensional configurations.
In recent years, the rise of deep neural networks has revolutionized various domains. The release of AlphaFold by DeepMind, which can accurately model protein structures from amino acid sequences, has spurred a surge in AI applications for protein structure prediction.
Video: Uncovering Protein Ensembles: Automated Multiconformer Model Building for Crystallography and CryoEM - YouTube
AlphaFold not only generates precise 3D predictions for protein monomers but has also expanded to include multimers through its AlphaFold-Multimer model.
Section 1.2: Innovations by Colossal-AI
To further enhance AlphaFold's capabilities, the Colossal-AI team recently introduced FastFold, an optimized and open-source version of AlphaFold. This implementation has successfully reduced training duration from 11 days to just 67 hours, while also accelerating inference times by approximately 11.6 times. Colossal-AI is committed to democratizing large-scale AI applications in the pharmaceutical industry.
Interactions between proteins are vital for their biological roles. To tackle the challenges associated with predicting the structures of monomers and multimers, Colossal-AI has unveiled the xTrimo Multimer model. This model better captures protein interactions, thus improving target analysis, protein structure simulation, and high-precision antibody designs crucial for drug discovery.
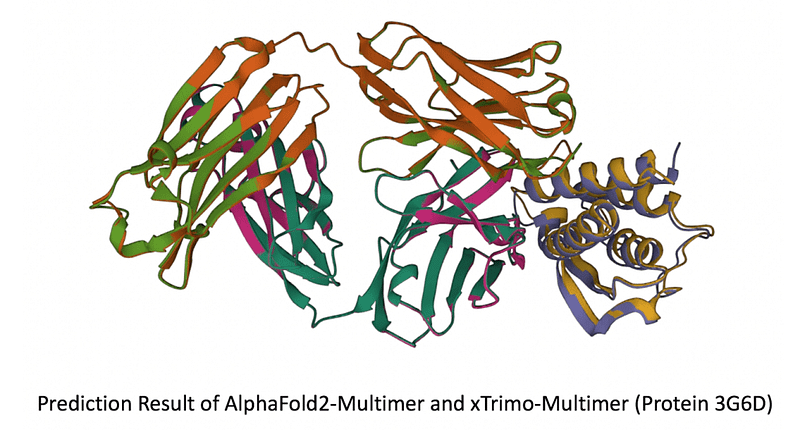
Chapter 2: Performance Enhancements of xTrimo Multimer
The high costs associated with AlphaFold's inference have posed challenges in its research and development, particularly with lengthy sequences that increase computational demands and memory usage. To address these issues, the Colossal-AI team has implemented CUDA optimization and Kernel Fusion techniques in xTrimo Multimer, resulting in significant performance improvements. The xTrimo Multimer outperforms both AlphaFold2 and OpenFold from Columbia University, achieving inference performance enhancements of 1.58 to 2.14 times and 1.14 to 2.23 times, respectively.
Video: Navigating the Structural Frontier with Protein NMR in the Era of Artificial Intelligence
Moreover, the xTrimo Multimer model is designed to support distributed inference for long sequences. By incorporating Dynamic Axial Parallelism, it effectively allocates computation and GPU memory across multiple devices, resolving the computational and memory challenges posed by lengthy sequences. The model achieves a speedup of 8.47 times and 11.15 times compared to OpenFold and AlphaFold2, respectively, for sequences ranging from 2,000 to 3,000 amino acids. Additionally, xTrimo Multimer can handle sequences up to 4,000 amino acids, a feat that OpenFold and AlphaFold2 cannot manage due to memory limitations. This allows scientists to complete a 4,000-length sequence inference in approximately 20 minutes.
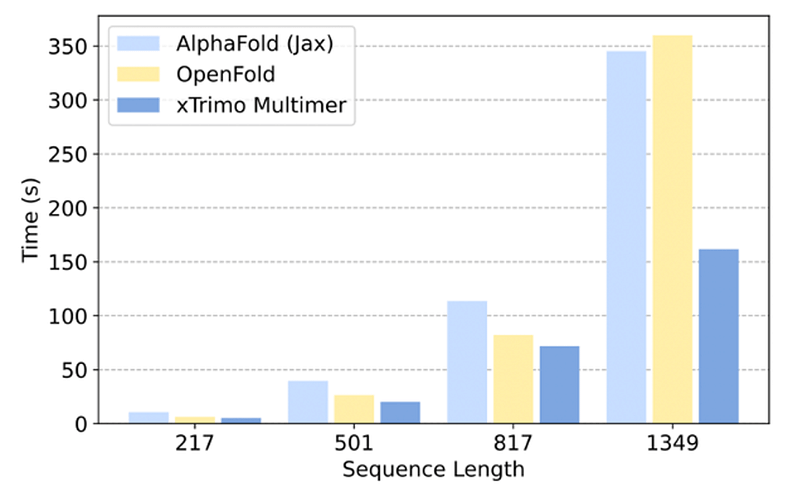
“The collaboration with HPC-AI Tech combines cutting-edge large AI model training technology from the Colossal-AI team with BioMap's expertise in biocomputing. The launch of xTrimo Multimer marks a significant milestone in integrating the benefits of large AI model training into BioMap’s xTrimo multimodal system,” stated Le Song, Chief AI Scientist at BioMap.
“Our latest protein monomer and multimer structure prediction solution represents a key advancement for Colossal-AI in addressing real-world industrial challenges. We aim to deepen our collaboration with BioMap in biocomputing large models, promoting the application of deep learning in innovative drug development,” remarked Yang You, Chairman of HPC-AI Tech.
The xTrimo Multimer serves as a vital product among other remarkable industrial solutions developed through Colossal-AI, facilitating large-scale AI modeling for global enterprises. The Colossal-AI team remains dedicated to exploring new possibilities in AI model training across various sectors, striving to address contemporary industry challenges and shaping the future of the global AI landscape.
Portal
About BioMap
BioMap consists of a team of esteemed scientists specializing in disease biology, bioinformatics, machine learning, and antibody engineering. Co-founded by Robin Li, CEO of Baidu, and Wei Liu, former CEO of Baidu Ventures, BioMap is dedicated to delivering first-in-class therapies for unmet medical needs in areas such as immune-oncology, autoimmune disorders, fibrosis, and age-related diseases.
About HPC-AI Tech
HPC-AI Tech is a global organization focused on enhancing the efficiency of training and deploying large AI models. Founded by Dr. Yang You, a Ph.D. graduate in Computer Science from UC Berkeley and currently a Presidential Young Professor at the National University of Singapore, HPC-AI Tech has developed an effective system for large AI model training and inference, Colossal-AI, which integrates state-of-the-art technologies to help users achieve efficient deployments at low costs.
We know you don’t want to miss any news or research breakthroughs. Subscribe to our popular newsletter Synced Global AI Weekly to get weekly AI updates.